Real-time PCR (qPCR) is a powerful technique for accurate analysis of gene expression. When starting with RNA samples, you first need to perform a reverse transcription (RT) step in order to generate cDNA for the qPCR experiment. In one-step RT-qPCR, the RT step takes place in the same tube as the qPCR. In two-step RT-qPCR, the RT and qPCR steps take place in separate tubes. Both techniques have advantages and limitations, so it is essential to consider which protocol suits your workflow.
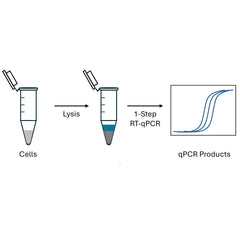
One-step RT-qPCR workflow
The single-tube protocol is easy to set up and can be used for processing many samples using liquid handlers or other high-throughput systems. Since both the RT and qPCR steps take place in the same tube, the reaction conditions cannot be optimized separately. This can lead to lower yields or efficiency in either step. Another limitation is that all the cDNA generated is used up in the subsequent qPCR step.

Two-step RT-qPCR workflow
The two-tube protocol makes it possible to optimize the RT and qPCR steps separately, ensuring maximum specificity and efficiency. This approach works best with workflows analyzing many targets in fewer samples. Another advantage is that two-step RT-qPCR generates cDNA in a separate tube from the qPCR reaction. This allows cDNA stocks to be banked so that they can be used for additional targets or validation. However, the protocols are more time-consuming and involve more pipetting steps. This introduces more variability and increases the risk of contamination. Secondly, the protocol cannot be adapted to automation easily.